Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans
Abstract
Machine learning methods offer great promise for fast and accurate detection and prognostication of coronavirus disease 2019 (COVID-19) from standard-of-care chest radiographs (CXR) and chest computed tomography (CT) images. Many articles have been published in 2020 describing new machine learning-based models for both of these tasks, but it is unclear which are of potential clinical utility. In this systematic review, we consider all published papers and preprints, for the period from 1 January 2020 to 3 October 2020, which describe new machine learning models for the diagnosis or prognosis of COVID-19 from CXR or CT images. All manuscripts uploaded to bioRxiv, medRxiv and arXiv along with all entries in EMBASE and MEDLINE in this timeframe are considered. Our search identified 2,212 studies, of which 415 were included after initial screening and, after quality screening, 62 studies were included in this systematic review. Our review finds that none of the models identified are of potential clinical use due to methodological flaws and/or underlying biases. This is a major weakness, given the urgency with which validated COVID-19 models are needed. To address this, we give many recommendations which, if followed, will solve these issues and lead to higher-quality model development and well-documented manuscripts.
Main
In December 2019, a novel coronavirus was first recognized in Wuhan, China1. On 30 January 2020, as infection rates and deaths across China soared and the first death outside China was recorded, the World Health Organization (WHO) described the then-unnamed disease as a Public Health Emergency of International Concern2. The disease was officially named coronavirus disease 2019 (COVID-19) by 11 February 20203, and was declared a pandemic on 11 March 20204. Since its first description in late 2019, the COVID-19 infection has spread across the globe, causing massive societal disruption and stretching our ability to deliver effective healthcare. This was caused by a lack of knowledge about the virus’s behaviour along with a lack of an effective vaccine and antiviral therapies.
Although PCR with reverse transcription (RT–PCR) is the test of choice for diagnosing COVID-19, imaging can complement its use to achieve greater diagnostic certainty or even be a surrogate in some countries where RT–PCR is not readily available. In some cases, chest radiograph (CXR) abnormalities are visible in patients who initially had a negative RT–PCR test5 and several studies have shown that chest computed tomography (CT) has a higher sensitivity for COVID-19 than RT–PCR, and could be considered as a primary tool for diagnosis6,7,8,9. In response to the pandemic, researchers have rushed to develop models using artificial intelligence (AI), in particular machine learning, to support clinicians.
Given recent developments in the application of machine learning models to medical imaging problems10,11, there is fantastic promise for applying machine learning methods to COVID-19 radiological imaging for improving the accuracy of diagnosis, compared with the gold-standard RT–PCR, while also providing valuable insight for prognostication of patient outcomes. These models have the potential to exploit the large amount of multimodal data collected from patients and could, if successful, transform detection, diagnosis and triage of patients with suspected COVID-19. Of greatest potential utility is a model that can not only distinguish patients with COVID-19 from patients without COVID-19 but also discern alternative types of pneumonia such as those of bacterial or other viral aetiologies. With no standardization, AI algorithms for COVID-19 have been developed with a very broad range of applications, data collection procedures and performance assessment metrics. Perhaps as a result, none are currently ready to be deployed clinically. Reasons for this include: (1) the bias in small datasets; (2) the variability of large internationally sourced datasets; (3) the poor integration of multistream data, particularly imaging data; (4) the difficulty of the task of prognostication; and (5) the necessity for clinicians and data analysts to work side-by-side to ensure the developed AI algorithms are clinically relevant and implementable into routine clinical care. Since the pandemic began in early 2020, researchers have answered the ‘call to arms’ and numerous machine learning models for diagnosis and prognosis of COVID-19 using radiological imaging have been developed and hundreds of manuscripts have been written. In this Analysis, we reviewed the entire literature of machine learning methods as applied to chest CT and CXR for the diagnosis and prognosis of COVID-19. As this is a rapidly developing field, we reviewed both published and preprint studies to ensure maximal coverage of the literature.
While earlier reviews provided a broad analysis of predictive models for COVID-19 diagnosis and prognosis12,13,14,15, this Analysis highlights the unique challenges researchers face when developing classical machine learning and deep learning models using imaging data. This Analysis builds on the approach of Wynants et al.12: we assess the risk of bias in the papers considered, going further by incorporating a quality screening stage to ensure only those papers with sufficiently documented methodologies are reviewed in most detail. We also focus our review on the systematic methodological flaws in the current machine learning literature for COVID-19 diagnosis and prognosis models using imaging data. We also give detailed recommendations in five domains: (1) considerations when collating COVID-19 imaging datasets that are to be made public; (2) methodological considerations for algorithm developers; (3) specific issues about reproducibility of the results in the literature; (4) considerations for authors to ensure sufficient documentation of methodologies in manuscripts; and (5) considerations for reviewers performing peer review of manuscripts.
This Analysis has been performed, and informed, by both clinicians and algorithm developers, with our recommendations aimed at ensuring the most clinically relevant questions are addressed appropriately, while maintaining standards of practice to help researchers develop useful models and report reliable results even in the midst of a pandemic.
Results
Study selection
Our initial search highlighted 2,212 papers that satisfied our search criteria; removing duplicates we retained 2,150 papers, and of these, 415 papers had abstracts or titles deemed relevant to the review question, introducing machine learning methods for COVID-19 diagnosis or prognosis using radiological imaging. Full-text screening retained 320 papers, of which, after quality review, 62 were included for discussion in this Analysis (Fig. 1). Of these, 37 were deep learning papers, 23 were traditional machine learning papers and 2 were hybrid papers (using both approaches).
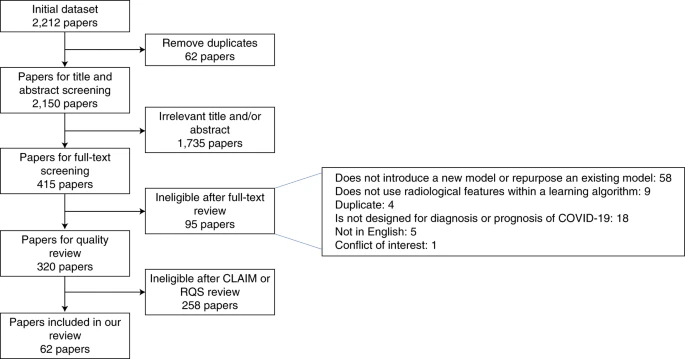
The flowchart highlights the inclusion and exclusion of papers at each stage.
Quality screening failures
Deep learning papers
There were 254/320 papers that described deep learning-based models and 215 of these were excluded from the detailed review (including one hybrid paper). We found that 110 papers (51%) fail at least three of our identified mandatory criteria from the CLAIM checklist (Supplementary Discussion 1), with 23% failing two and 26% failing just one. In the rejected papers, the three most common reasons for a paper failing the quality check was due to insufficient documentation of the following.
- (1)How the final model was selected in 61% (132)
- (2)The method of pre-processing of the images in 58% (125)
- (3)The details of the training approach (for example, the optimizer, the loss function, the learning rate) in 49% (105)
Traditional machine learning papers
There were 69 papers that described traditional machine learning methods and 44 of these were excluded from the review, that is, the RQS was less than 6 or the datasets used were not specified in the paper. There were only two papers that had an RQS ≥6, but which failed to disclose the datasets used in the analysis. Of the remaining papers, the two factors that lead to the lowest RQS results were omission of the following.
- (1)Feature reduction techniques in 52% of papers (23)
- (2)Model validation in 61% of papers (27)
The two hybrid papers both failed the checklist for artificial intelligence in medical imaging (CLAIM) check but passed the radiomic quality score (RQS) criteria. Full details can be found in Supplementary Data 2.
Remaining papers for detailed analysis
Deep learning papers
There were six non-mandatory CLAIM criteria not satisfied in at least half of the 37 papers.
- (1)29 did not complete any external validation
- (2)30 did not perform any robustness or sensitivity analysis of their model
- (3)26 did not report the demographics of their data partitions
- (4)25 did not report the statistical tests used to assess significance of results or determine confidence intervals
- (5)23 did not report confidence intervals for the performance
- (6)22 did not sufficiently report their limitations, biases or issues around generalizability
The full CLAIM results are in Supplementary Data 2.
Traditional machine learning papers
Of the 25 papers, including the two hybrid papers, none used longitudinal imaging, performed a prospective study for validation, or standardized image acquisition by using either a phantom study or a public protocol. Only six papers described performing external validation and only four papers reported the calibration statistics (the level of agreement between predicted risks and those observed) and associated statistical significance for the model predictions. The full RQS scores are in Supplementary Data 2.
Datasets considered
Public datasets were used extensively in the literature appearing in 32/62 papers (see Supplementary Discussion 2 for list of public datasets, three papers use both public and private data). Private data were used in 33/62 papers with 21 using data from mainland China, three using data from France and the remainder using data from Iran, the United States, Belgium, Brazil, Hong Kong and the Netherlands.
Diagnostic models for COVID-19
Diagnosis models using CXRs
Twenty-two papers considered diagnosis of COVID-19 from CXR images16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36. Most of these papers used off-the-shelf networks, including ResNet-18 or ResNet-5016,17,20,26,29,32,37, DenseNet-12127,28,31,32,34, VGG-16 or VGG-1919,33,35, Inception21,38 and EfficientNet30,39, with three considering custom architectures18,25,36 and three using hand-engineered features22,23,24. Most papers classified images into the three classes, that is, COVID-19, non-COVID-19 pneumonia and normal16,19,21,23,25,26,28,30,32,33,34,35,36,37, while two considered an extra class by dividing non-COVID-19 pneumonia into viral and bacterial pneumonia17,29. ResNet and DenseNet architectures showed better performance than the others, with accuracies ranging from 0.88 to 0.99. However, we caution against direct comparison as the papers use different training and testing settings (for example, different datasets and data partition sizes) and consider a different number of classes.
Diagnostic models using CT scans and deep learning
Eighteen papers applied deep learning techniques to CT imaging, all of which were framed as a classification task to distinguish COVID-19 from other lung pathologies such as (viral or bacterial) pneumonia, interstitial lung disease35,40,41,42,43,44,45,46,47 and/or a non-COVID-19 class40,41,44,46,48,49,50,51,52. The full three-dimensional (3D) volumes were only considered in seven papers40,43,47,50,52,53,54 with the remainder considering isolated 2D slices or even 2D patches45. In most 2D models, authors employed transfer learning, with networks pre-trained on ImageNet55. Almost all models used lung segmentation as a pre-processing step. One paper48 used a generative adversarial network56 approach to address the paucity of COVID-19 CT imaging. Values reported for the area under the receiver operating characteristic curve (AUC) ranged from 0.70 to 1.00.
Diagnostic models using CT scans and traditional machine learning methods
Eight papers employed traditional machine learning methods for COVID-19 diagnosis using hand-engineered features40,57,58,59,60,61,62 or convolutional neural network (CNN)-extracted features46. Four papers46,59,60,62 incorporated clinical features with those obtained from the CT images. All papers using hand-engineered features employed feature reduction, using between 4 and 39 features in their final models. For final classification, five papers used logistic regression40,58,59,60,61, one used a random forest57, one a multilayer perceptron46 and one compared many different machine learning classifiers to determine the best62. Accuracies ranged from 0.76 to 0.98 (refs. 40,46,57,58,59). As before, we caution against direct comparison. The traditional machine-learning model in the hybrid paper40 had a 0.05 lower accuracy than their deep learning model.
Prognostic models for COVID-19 using CT and CXR images
Nineteen papers developed models for the prognosis of patients with COVID-1951,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80, fifteen using CT and four using CXR. These models were developed for predicting severity of outcomes including: death or need for ventilation72,78,79, a need for intensive care unit (ICU) admission63,73,77,78,79, progression to acute respiratory distress syndrome80, the length of hospital stay51,74, likelihood of conversion to severe disease64,65,75 and the extent of lung infection76. Most papers used models based on a multivariable Cox proportional hazards model51,72,78,79, logistic regression65,73,74,75,80, linear regression75,76, random forest74,77 or compare a huge variety of machine learning models such as tree-based methods, support vector machines, neural networks and nearest-neighbour clustering63,64.
Predictors from radiological data were extracted using either handcrafted radiomic features63,64,68,69,70,72,74,75,77,78,79,80 or deep learning51,66,70,71,73,76. Clinical data included basic observations, serology and comorbidities. Only eight models integrated both radiological and clinical data62,63,69,72,73,77,78,79.
Risks of bias
Following the prediction model risk of bias assessment tool (PROBAST) guidance, the risk of bias was assessed for all 62 papers in four domains: participants, predictors, outcomes and analysis. The results are shown in Table 1. We found that 55/62 papers had a high risk of bias in at least one domain with the others unclear in at least one domain.
Participants
Almost all papers had a high (45/62) or unclear (11/62) risk of bias for their participants, with only six assessed as having a low risk of bias. This was primarily due to the following issues: (1) for public datasets it is not possible to know whether patients are truly COVID-19 positive, or if they have underlying selection biases, as anybody can contribute images16,24,26,28,29,30,31,32,34,35,37,41,44,48,49,76; (2) the paper uses only a subset of original datasets, applying some exclusion criteria, without enough details to be reproducible16,43,44,48,49,51,61,70,71,75,76; and/or (3) there are large differences in demographics between the COVID-19 cohort and the control groups, with, for example, paediatric patients as controls17,24,28,29,31,32,35,37,45,46,59,81.
Predictors
For models where the features have been extracted using deep learning models, the predictors are unknown and abstract imaging features. Therefore, for these papers (38/62), we could not judge biases in the predictors. For 20 papers, the risk of bias was recorded as low due to the use of pre-defined hand-engineered features. For the remaining 4 papers, a high risk of bias was recorded due to the predictors being assessed with knowledge of the associated outcome.
Outcomes
The risk of bias in the outcome variable was found to be low for the majority (25/62) of the papers, unclear for 26/62 and high for 11/62. To evaluate the bias in the outcome, we took different approaches for papers using private datasets and public datasets (three papers use a mixture).
For the 35 papers that used public datasets, the outcome was assigned by the originators of the dataset and not by the papers’ authors. Papers that used a public dataset generally have an unclear risk of bias (30/35) as they used the outcome directly sourced from the dataset originator.
For the 33 papers that used private datasets, the COVID-19 diagnosis was due to either positive RT–PCR or antibody tests for 24/33 and have a low risk of bias. The other papers have a high (7/33) or unclear (2/33) risk of bias due to inconsistent diagnosis of COVID-1940,82, unclear definition of a control group63,65, ground truths being assigned using the images themselves26,54,60,71, using an unestablished reference to define outcome74 or by combining public and private datasets41,66,83.
Analysis
Only ten papers have a low risk of bias for their analysis. The high risk of bias in most papers was principally due to a small sample size of patients with COVID-19 (leading to highly imbalanced datasets), use of only a single internal holdout set for validating their algorithm (rather than cross-validation or bootstrapping) and a lack of appropriate evaluation of the performance metrics (for example, no discussion of calibration/discrimination)18,19,20,22,23,44,48,52,64,72,80. One paper with a high risk of bias32 claimed external validation on the dataset from ref. 84, not realizing that this already includes datasets from both ref. 85 and ref. 86 that were used to train the algorithm.
Data analysis
There are two approaches for validating the performance of an algorithm, namely internal and external validation. For internal validation, the test data are from the same source as the development data and for external validation they are from different sources. Including both internal and external validation allows more insight to generalizability of the algorithm. We found that 48/62 papers consider internal validation only, with 13/62 using external validation22,32,41,42,51,54,63,66,67,69,73,78,79. Twelve used truly external test datasets and one tested on the same data the algorithm was trained on32.
Model evaluation
In Table 2, we give the performance metrics quoted in each paper. Ten papers used cross-validation to evaluate model performance21,35,36,47,49,57,65,72,74,75,77, one used both cross-validation and an external test set41, one quoted correlation metrics76 and one had an unclear validation method17. The other papers all had an internal holdout or external test set with sensitivity and specificity derived from the test data using an unquoted operating point (with the exception of ref. 16, which quotes operating point 0.5). It would be expected that an operating point be chosen based on the algorithm performance for the validation data used to tune and select the final algorithm. However, the receiver operating characteristic (ROC) curves and AUC values are given for the internal holdout or external test data independent of the validation data.
Partition analysis
In Fig. 2, we show the quantity of data (split by class) used in the training cohort of 32 diagnosis models. We excluded many studies18,20,22,23,25,28,29,32,35,43,45,58,71 because it was unclear how many images were used. If a paper only stated the number of patients (and not the number of images), we assumed that there was only one image per patient. We see that 20/32 papers have a reasonable balance between classes (with exceptions being refs. 17,24,26,30,31,33,36,37,40,51,61,62. However, the majority of datasets were quite small, with 19/32 papers using fewer than 2,000 datapoints for development (with exceptions being refs. 17,18,26,27,30,31,33,36,41,48,53,57,81). Only seven papers used both a dataset with more than 2,000 datapoints that was balanced for COVID-19 positive and the other classes27,41,48,53,54,57,81.
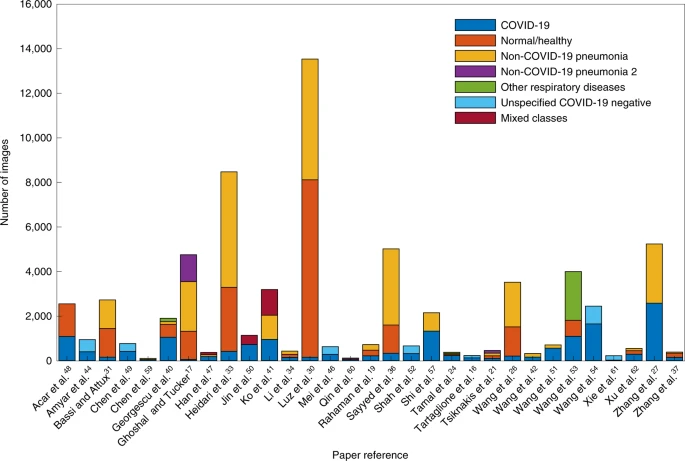
It is noted that we exclude Bai et al.81 from the figure as they used much more training data (118,401 images) than other papers. For Xu et al.62, two COVID-19 classes are shown in the graph as one combined class.
Figure 3 shows the number of images of each class used in the holdout/test cohorts. We found that 6/32 papers had an imbalanced testing dataset17,24,33,36,37,61. Only 6/32 papers tested on more than 1,000 images17,27,36,41,54,81. Only 4/32 had both a large and balanced testing dataset27,41,54,81.
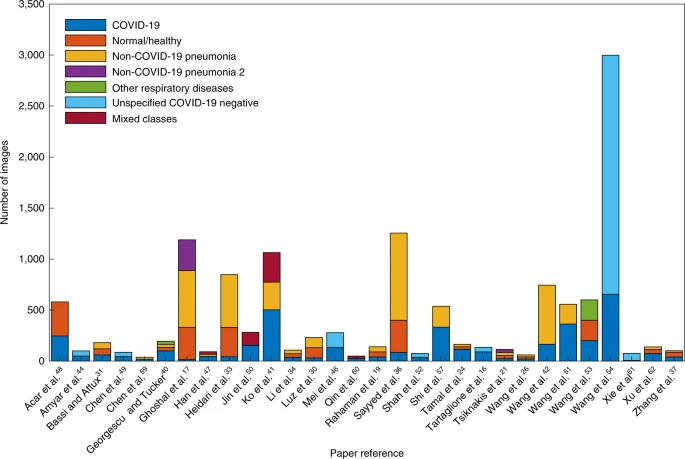
It is noted that we exclude Bai et al.81 and Zhang et al.27 from the figure as they used far more testing data (14,182 and 5,869 images respectively) than other papers. There were a large number of images (1,237) in the testing dataset in Wang et al.54 that were unidentified in the paper (we include these in the unspecified COVID-19 negative).
Public availability of the algorithms and models
Only 13/62 papers21,23,27,30,34,36,46,51,66,67,74,76,81 published the code for reproducing their results (seven including their pre-trained parameters) and one stated that it is available on request74.
Discussion
Our systematic review highlights the extensive efforts of the international community to tackle the COVID-19 pandemic using machine learning. These early studies show promise for diagnosis and prognostication of pneumonia secondary to COVID-19. However, we have also found that current reports suffer from a high prevalence of deficiencies in methodology and reporting, with none of the reviewed literature reaching the threshold of robustness and reproducibility essential to support utilization in clinical practice. Many studies are hampered by issues with poor-quality data, poor application of machine learning methodology, poor reproducibility and biases in study design. The current paper complements the work of Wynants et al. who have published a living systematic review12 on publications and preprints of studies describing multivariable models for screening of COVID-19 infections in the general population, differential diagnosis of COVID-19 infection in patients that are symptomatic and prognostication in patients with confirmed COVID-19 infection. While Wynants et al. reviewed multivariable models with any type of clinical input data, the present review focuses specifically on machine learning-based diagnostic and prognostic models using medical imaging. Furthermore, this systematic review employed specialized quality metrics for the assessment of radiomics and deep learning-based diagnostic models in radiology. This is also in contrast to previous studies that have assessed AI algorithms in COVID-1913,14. Limitations of the current literature most frequently reflect either a limitation of the dataset used in the model or methodological mistakes repeated in many studies that probably lead to overly optimistic performance evaluations.
Datasets
Many papers gave little attention to establishing the original source of the images (Supplementary Discussion 2). When considering papers that use public data, readers should be aware of the following.
Duplication and quality issues
There is no restriction for a contributor to upload COVID-19 images to many of the public repositories85,87,88,89,90. There is high likelihood of duplication of images across these sources and no assurance that the cases included in these datasets are confirmed COVID-19 cases (authors take a great leap to assume this is true) so great care must be taken when combining datasets from different public repositories. Also, most of the images have been pre-processed and compressed into non-DICOM formats leading to a loss in quality and a lack of consistency/comparability.
- Source issues. Many papers (16/62) used the pneumonia dataset of Kermany et al.86 as a control group. They commonly failed to mention that this consists of paediatric patients aged between one and five. Developing a model using adult patients with COVID-19 and very young patients with pneumonia is likely to overperform as it is merely detecting children versus adults. This dataset is also erroneously referred to as the Mooney dataset in many papers (being the Kermany dataset deployed on Kaggle91). It is also important to consider the sources of each image class, for example, if images for different diagnoses are from different sources. It is demonstrated by Maguolo et al.92 that by excluding the lung region entirely, the authors could identify the source of the images in the Cohen et al.85 and Kermany et al.86 datasets with an AUC between 0.9210 and 0.9997, and ‘diagnose’ COVID-19 with an AUC = 0.68.
- Frankenstein datasets. The issues of duplication and source become compounded when public ‘Frankenstein’ datasets are used, that is, datasets assembled from other datasets and redistributed under a new name. For instance, one dataset91 combined several other datasets85,88,93 without realizing that one of the component datasets93 already contains another component88. This repackaging of datasets, although pragmatic, inevitably leads to problems with algorithms being trained and tested on identical or overlapping datasets while believing them to be from distinct sources.
- Implicit biases in the source data. Images uploaded to a public repository and those extracted from publications93 are likely to have implicit biases due to the contribution source. For example, it is likely that more interesting, unusual or severe cases of COVID-19 appear in publications.
Methodology
All proposed models suffer from a high or unclear risk of bias in at least one domain. There are several methodological issues driven by the urgency in responding to the COVID-19 crisis and subtler sources of bias due to poor application of machine learning.
The urgency of the pandemic led to many studies using datasets that contain obvious biases or are not representative of the target population, for example, paediatric patients. Before evaluating a model, it is crucial that authors report the demographic statistics for their datasets, including age and sex distributions. Diagnostic studies commonly compare their models’ performance to that of RT–PCR. However, as the ground-truth labels are often determined by RT–PCR, there is no way to measure whether a model outperforms RT–PCR from accuracy, sensitivity or specificity metrics alone. Ideally, models should aim to match clinicians using all available clinical and radiomic data, or to aid them in decision-making.
Many papers utilized transfer learning in developing their model, which assumes an inherent benefit to performance. However, it is unclear whether transfer learning offers a large performance benefit due to the over-parameterization of the models41,58. Many publications used the same resolutions such as 224-by-224 or 256-by-256 for training, which are often used for ImageNet classification, indicating that the pre-trained model dictated the image rescaling used rather than clinical judgement.
Recommendations
Based on the systematic issues we encountered in the literature, we offer recommendations in five distinct areas: (1) the data used for model development and common pitfalls; (2) the evaluation of trained models; (3) reproducibility; (4) documentation in manuscripts; and (5) the peer-review process. Our recommendations in areas (3) and (4) are largely informed by the 258 papers that did not pass our initial quality check, while areas (1), (2) and (5) follow from our analysis of the 62 papers receiving our full review.
Recommendations for data
First, we advise caution over the use of public repositories, which can lead to high risks of bias due to source issues and Frankenstein datasets as discussed above. Furthermore, authors should aim to match demographics across cohorts, an often neglected but important potential source of bias; this can be impossible with public datasets that do not include demographic information, and including paediatric images86 in the COVID-19 context introduces a strong bias.
Using a public dataset alone without additional new data can lead to community-wide overfitting on this dataset. Even if each individual study observes sufficient precautions to avoid overfitting, the fact that the community is focused on outperforming benchmarks on a single public dataset encourages overfitting. Many public datasets containing images taken from preprints receive these images in low-resolution or compressed formats (for example, JPEG and PNG), rather than their original DICOM format. This loss of resolution is a serious concern for traditional machine learning models if the loss of resolution is not uniform across classes, and the lack of DICOM metadata does not allow exploration of model dependence on image acquisition parameters (for example, scanner manufacturer, slice thickness and so on).
Regarding CXRs, researchers should be aware that algorithms might associate more severe disease not with CXR imaging features, but the view that has been used to acquire that CXR. For example, for patients that are sick and immobile, an anteroposterior CXR view is used for practicality rather than the standard posteroanterior CXR projection. Also, overrepresentation of severe disease is bad not only from the machine learning perspective but also in terms of clinical utility, as the most useful algorithms are those that can diagnose disease at an early stage94. The timing between imaging and RT–PCR tests was also largely undocumented, which has implications for the validity of the ground truth used. It is also important to recognize that a negative RT–PCR test does not necessarily mean that a patient does not have COVID-19. We encourage authors to evaluate their algorithms on datasets from the pre-COVID-19 era, such as performed by ref. 95, to validate any claims that the algorithm is isolating COVID-19-specific imaging features. It is common for non-COVID-19 diagnoses (for example, non-COVID-19 pneumonia) to be determined from imaging alone. However, in many cases, these images are the only predictors of the developed model, and using predictors to inform outcomes leads to optimistic performance.
Recommendations for evaluation
We emphasize the importance of using a well-curated external validation dataset of appropriate size to assess generalizability to other cohorts. Any useful model for diagnosis or prognostication must be robust enough to give reliable results for any sample from the target population rather than just on the sampled population. Calibration statistics should be calculated for the developed models to inform predictive error and decision curve analysis96 performed for assessing clinical utility. It is important for authors to state how they ensured that images from the same patient were not included in the different dataset partitions, such as describing patient-level splits. This is an issue for approaches that consider 2D and 3D images as a single sample and also for those that process 3D volumes as independent 2D samples. It is also important when using datasets containing multiple images from each patient. When reporting results, it is important to include confidence intervals to reflect the uncertainty in the estimate, especially when training models on the small sample sizes commonly seen with COVID-19 data. Moreover, we stress the importance of not only reporting results but also demonstrating model interpretability with methods such as saliency maps, which is a necessary consideration for adoption into clinical practice. We remind authors that it is inappropriate to compare model performance to RT–PCR or any other ground truths. Instead, authors should aim for models to either improve the performance and efficiency of clinicians, or, even better, to aid clinicians by providing interpretable predictions. Examples of interpretability techniques include: (1) informing the clinician of which features in the data most influenced the prediction of the model, (2) linking the prognostic features to the underlying biology and (3) overlaying an activation/saliency map on the image to indicate the region of the image that influenced the model’s prediction, and (4) identifying patients that had a similar clinical pathway.
Most papers derive their performance metrics from the test data alone with an unstated operating point to calculate sensitivity and specificity. Clinical judgement should be used to identify the desired sensitivity or specificity of the model and the operating point should be derived from the development data. The differences in the sensitivity and specificity of the model should be recorded separately for the validation and test data. Using an operating point of 0.5 and only reporting the test sensitivity and specificity fails to convey the reliability of the threshold. This is a key aspect of generalizability. Omitting it, in the process of device regulation, would see a US Food and Drug Administration 510K submission rejected.
Recommendations for replicability
A possible ambiguity arises due to updating of publicly available datasets or code. Therefore, we recommend that a cached version of the public dataset be saved, or the date/version quoted, and specific versions of data or code be appropriately referenced. (Git commit ids or tags can be helpful for this purpose to reference a specific version on GitHub, for example.) We acknowledge that although perfect replication is potentially not possible, details such as the seeds used for randomness and the actual partitions of the dataset for training, validation and testing would form very useful supplementary materials.
Recommendations for authors
For authors, we recommend assessing their paper against appropriate established frameworks, such as RQS, CLAIM, transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD), PROBAST and Quality Assessment of Diagnostic Accuracy Studies (QUADAS)97,98,99,100,101. By far the most common point leading to exclusion was failure to state the data pre-processing techniques in sufficient detail. As a minimum, we expected papers to state any image resizing, cropping and normalization used before model input, and with this small addition many more papers would have passed through the quality review stage. Other commonly missed points include details of the training (such as number of epochs and stopping criteria), robustness or sensitivity analysis, and the demographic or clinical characteristics of patients in each partition.
Recommendations for reviewers
For reviewers, we also recommend the use of the checklists97,98,99,100,101 to better identify common weaknesses in reporting the methodology. The most common issues in the papers we reviewed was the use of biased datasets and/or methodologies. For non-public datasets, it may be difficult for reviewers to assess possible biases if an insufficiently detailed description is given by the authors. We strongly encourage reviewers to ask for clarification from the authors if there is any doubt about bias in the model being considered. Finally, we suggest using reviewers from a combination of both medical and machine learning backgrounds, as they can judge the clinical and technical aspects in different ways.
Challenges and opportunities
Models developed for diagnosis and prognostication from radiological imaging data are limited by the quality of their training data. While many public datasets exist for researchers to train deep learning models for these purposes, we have determined that these datasets are not large enough, or of suitable quality, to train reliable models, and all studies using publicly available datasets exhibit a high or unclear risk of bias. However, the size and quality of these datasets can be continuously improved if researchers worldwide submit their data for public review. Because of the uncertain quality of many COVID-19 datasets, it is likely more beneficial to the research community to establish a database that has a systematic review of submitted data than it is to immediately release data of questionable quality as a public database.
The intricate link of any AI algorithm for detection, diagnosis or prognosis of COVID-19 infections to a clear clinical need is essential for successful translation. As such, complementary computational and clinical expertise, in conjunction with high-quality healthcare data, are required for the development of AI algorithms. Meaningful evaluation of an algorithm’s performance is most likely to occur in a prospective clinical setting. Like the need for collaborative development of AI algorithms, the complementary perspectives of experts in machine learning and academic medicine were critical in conducting this systematic review.
Limitations
Due to the fast development of diagnostic and prognostic AI algorithms for COVID-19, at the time of finalizing our analyses, several new preprints have been released; these are not included in this study.
Our study has limitations in terms of methodologic quality and exclusion. Several high-quality papers published in high-impact journals—including Radiology, Cell and IEEE Transactions on Medical Imaging—were excluded due to the lack of documentation on the proposed algorithmic approaches. As the AI algorithms are the core for the diagnosis and prognosis of COVID-19, we only included works that are reproducible. Furthermore, we acknowledge that the CLAIM requirements are harder to fulfil than the RQS ones, and the paper quality check is therefore not be fully comparable between the two. We underline that several excluded papers were preprint versions and may possibly pass the systematic evaluation in a future revision.
In our PROBAST assessment, for the ‘Were there a reasonable number of participants?’ question of the analysis domain, we required a model to be trained on at least 20 events per variable for the size of the dataset to score a low risk of bias100. However, events per variable may not be a useful metric to determine whether a deep learning model will overfit. Despite their gross over-parameterization, deep learning models generalize well in a variety of tasks, and it is difficult to determine a priori whether a model will overfit given the number of training examples102. A model that was trained using fewer than 500 COVID-19 positive images was deemed to have a high risk of bias in answer to this and more than 2,000 COVID-19 positive images qualified as low risk. However, in determining the overall risk of bias for the analysis domain, we factor in nine PROBAST questions, so it is possible for a paper using fewer than 500 images to achieve at best an unclear overall risk of bias for its analysis. Similarly, it is possible for papers that have over 2,000 images to have an overall high risk of bias for their analysis if it does not account for other sources of bias.
Conclusions
This systematic review specifically considers the current machine learning literature using CT and CXR imaging for COVID-19 diagnosis and prognosis, which emphasizes the quality of the methodologies applied and the reproducibility of the methods. We found that no papers in the literature currently have all of: (1) a sufficiently documented manuscript describing a reproducible method; (2) a method that follows best practice for developing a machine learning model; and (3) sufficient external validation to justify the wider applicability of the method. We give detailed specific recommendations for data curators, machine learning researchers, manuscript authors and reviewers to ensure the best-quality methods are developed that are reproducible and free from biases in either the underlying data or the model development.
Despite the huge efforts of researchers to develop machine learning models for COVID-19 diagnosis and prognosis, we found methodological flaws and many biases throughout the literature, leading to highly optimistic reported performance. In their current reported form, none of the machine learning models included in this review are likely candidates for clinical translation for the diagnosis/prognosis of COVID-19. Higher-quality datasets, manuscripts with sufficient documentation to be reproducible and external validation are required to increase the likelihood of models being taken forward and integrated into future clinical trials to establish independent technical and clinical validation as well as cost-effectiveness.
Methods
The methods for performing this systematic review are registered with PROSPERO (CRD42020188887) and were agreed by all authors before the start of the review process, to avoid bias.
Search strategy and selection criteria
We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklist103 and include this in Supplementary Table 1. We performed our search to identify published and unpublished works using the arXiv and the ‘Living Evidence on COVID-19’ database104, a collation of all COVID-19 related papers from EMBASE via OVID, MEDLINE via PubMed, bioRxiv and medRxiv. The databases were searched from 1 January 2020 to 3 October 2020. The full search strategy is detailed in ‘Search strategy’. The initial cut-off is chosen to specifically include all early COVID-19 research, given that the WHO was only informed of the ‘pneumonia of unknown cause’ on 31 December 2019105. An initial search was performed on 28 May 2020, with updated searches performed on 24 June 2020, 14 August 2020, 15 August 2020 and 3 October 2020 to identify any relevant new papers published in the intervening period. As many of the papers identified are preprints, some of them were updated or published between these dates; in such cases, we used the preprint as it was at the later search date or the published version. Some papers were identified as duplicates ourselves or by Covidence106; in these instances, we ensured that the latest version of the paper was reviewed. We used a three-stage process to determine which papers would be included in this review. During the course of the review, one author (A.I.A.-R.) submitted a paper107 that was in scope for this review; however, we excluded it due to the potential for conflict of interest.
Title and abstract screening
In the first stage, a team of ten reviewers assessed papers for eligibility, screening the titles and abstracts to ensure relevance. Each paper was assessed by two reviewers independently and conflicts were resolved by consensus of the ten reviewers (Supplementary Data 1).
Full-text screening
In the second stage, the full text of each paper was screened by two reviewers independently to ensure that the paper was eligible for inclusion with conflicts resolved by consensus of the ten reviewers.
Quality review
In the third stage, we considered the quality of the documentation of methodologies in the papers. Note that exclusion at this stage is not a judgement on the quality or impact of a paper or algorithm, merely that the methodology is not documented with enough detail to allow the results to be reliably reproduced.
At this point, we separated machine learning methods into deep learning methods and non-deep learning methods (we refer to these as traditional machine learning methods). The traditional machine learning papers were scored using the RQS of Lambin et al.97, while the deep learning papers were assessed against the CLAIM of Mongan et al.98. The ten reviewers were assigned to five teams of two: four of the ten reviewers have a clinical background and were paired with non-clinicians in four of the five teams to ensure a breadth of experience when reviewing these papers. Within each team, the two reviewers independently assessed each paper against the appropriate quality measure. Where papers contained both deep learning and traditional machine learning methodologies, these were assessed using both CLAIM and RQS. Conflicts were resolved by a third reviewer.
To restrict consideration to only those papers with the highest-quality documentation of methodology, we excluded papers that did not fulfil particular CLAIM or RQS requirements. For the deep learning papers evaluated using the CLAIM checklist, we selected eight checkpoint items deemed mandatory to allow reproduction of the paper’s method and results. For the traditional machine learning papers, evaluated using the RQS, we used a threshold of 6 points out of 36 for inclusion in the review along with some basic restrictions, such as detail of the data source and how subsets were selected. The rationale for these CLAIM and RQS restrictions is given in Supplementary Discussion 1. If a paper was assessed using both CLAIM and RQS then it only needed to pass one of the quality checks to be included.
In a number of cases, various details of pre-processing, model configuration or training setup were not discussed in the paper, even though they could be inferred from a referenced online code repository (typically GitHub). In these cases, we have assessed the papers purely on the content in the paper, as it is important to be able to reproduce the method and results independently of the authors’ code.
Risk of bias in individual studies
We use the PROBAST of Wolff et al.100 to assess bias in the datasets, predictors and model analysis in each paper. The papers that passed the quality assessment stage were split among three teams of two reviewers to complete the PROBAST review. Within each team, the two reviewers independently scored the risk of bias for each paper and then resolved by conflicts any remaining conflicts were resolved by a third reviewer.
Data analysis
The papers were allocated among five teams of two reviewers. These reviewers independently extracted the following information: (1) whether the paper described a diagnosis or prognosis model; (2) the data used to construct the model; (3) whether there were predictive features used for the model construction; (4) the sample sizes used for the development and holdout cohorts (along with the number of COVID-19 positive cases); (5) the type of validation performed; (6) the best performance quoted in the paper for the validation cohort (whether internal, external or both); and (7) whether the code for training the model and the trained model were publicly available. Any conflicts were initially resolved by team discussions and remaining conflicts were resolved by a third reviewer.
Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation or writing of the manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Search strategy
Initial extraction
For the arXiv papers, we initially extracted papers for the relevant date ranges that included ‘ncov’ (as a complete word), ‘coronavirus’, ‘covid’, ‘sars-cov-2’ or ‘sars-cov2’ in their title or abstract. For the ‘Living Evidence on COVID-19’ database104, we downloaded all papers in the appropriate date range.
Refined search
We then filtered the identified papers using the following criteria: title or abstract contain one of: ‘ai’, ’deep’, ’learning’, ‘machine’, ‘neural’, ‘intelligence’, ‘prognos’, ‘diagnos’, ‘classification’, ‘ segmentation’ and also contain one of ‘ct’, ‘cxr’, ‘x-ray’, ‘xray’, ‘imaging’, ‘image’, ‘radiograph’. Only ‘ai’, ‘ct’ and ‘cxr’ are required to be complete words. This differs slightly from the original PROSPERO description; the search was widened to identify some additional papers. The full history of the searching and filtering source code can be explored at https://gitlab.developers.cam.ac.uk/maths/cia/covid-19-systematic-review
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files).
References
-
Wang, D. et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323, 1061–1069 (2020).
-
Zheng, Y. Y., Ma, Y. T., Zhang, J. Y. & Xie, X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 17, 259–260 (2020).
-
WHO Director-General’s Remarks at the Media Briefing on 2019-nCoV on 11 February 2020 (World Health Organization, 2020); https://www.who.int/director-general/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020
-
WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020 (World Health Organization, 2020); https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020
-
Wong, H. Y. F. et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology 296, 201160 (2019).
-
Long, C. et al. Diagnosis of the coronavirus disease (COVID-19): rRT–PCR or CT? Eur. J. Radiol. 126, 108961 (2020).
-
Fang, Y. et al. Sensitivity of chest CT for COVID-19: comparison to RT–PCR. Radiology 296, 200432 (2020).
-
Ai, T. et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology https://doi.org/10.1148/radiol.2020200642 (2020).
-
Sperrin, M., Grant, S. W. & Peek, N. Prediction models for diagnosis and prognosis in COVID-19. BMJ 369, m1464 (2020).
-
Irvin, J. et al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. Proc. AAAI Conf. Artif. Intell. 33, 590–597 (2019).
-
Huang, P. et al. Prediction of lung cancer risk at follow-up screening with low-dose CT: a training and validation study of a deep learning method. Lancet Digit. Health 1, e353–e362 (2019).
-
Wynants, L. et al. Prediction models for diagnosis and prognosis of COVID-19: systematic review and critical appraisal. BMJ 369, m1328 (2020).
-
Hamzeh, A. et al. Artificial intelligence techniques for containment COVID-19 pandemic: a systematic review. Res. Sq. https://doi.org/10.21203/rs.3.rs-30432/v1 (2020).
-
Albahri, O. S. et al. Systematic review of artificial intelligence techniques in the detection and classification of COVID-19 medical images in terms of evaluation and benchmarking: taxonomy analysis, challenges, future solutions and methodological aspects. J. Infect. Public Health https://doi.org/10.1016/j.jiph.2020.06.028 (2020).
-
Feng, S. et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 14, 4–15 (2021).
-
Tartaglione, E., Barbano, C. A., Berzovini, C., Calandri, M. & Grangetto, M. Unveiling COVID-19 from chest X-ray with deep learning: a hurdles race with small data. Int. J. Environ. Res. Public Health 17, 6933 (2020).
-
Ghoshal, B. & Tucker, A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. Preprint at http://arxiv.org/abs/2003.10769 (2020).
-
Malhotra, A. et al. Multi-task driven explainable diagnosis of COVID-19 using chest X-ray images. Preprint at https://arxiv.org/abs/2008.03205 (2020).
-
Rahaman, M. M. et al. Identification of COVID-19 samples from chest X-ray images using deep learning: a comparison of transfer learning approaches. J. Xray. Sci. Technol. 28, 821–839 (2020).
-
Amer, R., Frid-Adar, M., Gozes, O., Nassar, J. & Greenspan, H. COVID-19 in CXR: from detection and severity scoring to patient disease monitoring. Preprint at https://arxiv.org/abs/2008.02150 (2020).
-
Tsiknakis, N. et al. Interpretable artificial intelligence framework for COVID‑19 screening on chest X‑rays. Exp. Ther. Med. 20, 727–735 (2020).
-
Elaziz, M. A. et al. New machine learning method for imagebased diagnosis of COVID-19. PLoS ONE 15, e0235187 (2020).
-
Gil, D., Díaz-Chito, K., Sánchez, C. & Hernández-Sabaté, A. Early screening of SARS-CoV-2 by intelligent analysis of X-ray images. Preprint at https://arxiv.org/abs/2005.13928 (2020).
-
Tamal, M. et al. An integrated framework with machine learning and radiomics for accurate and rapid early diagnosis of COVID-19 from chest X-ray. Preprint at medRxiv https://doi.org/10.1101/2020.10.01.20205146 (2020).
-
Bararia, A., Ghosh, A., Bose, C. & Bhar, D. Network for subclinical prognostication of COVID 19 patients from data of thoracic roentgenogram: a feasible alternative screening technology. Preprint at medRxiv https://doi.org/10.1101/2020.09.07.20189852 (2020).
-
Wang, Z. et al. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recognit. 110, 107613 (2021).
-
Zhang, R. et al. Diagnosis of COVID-19 pneumonia using chest radiography: value of artificial intelligence. Radiology https://doi.org/10.1148/radiol.2020202944 (2020).
-
Ezzat, D., Hassanien, A. E. & Ella, H. A. An optimized deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization. Appl. Soft Comput. J. https://doi.org/10.1016/j.asoc.2020.106742 (2020).
-
Farooq, M. & Hafeez, A. COVID-ResNet: a deep learning framework for screening of COVID19 from radiographs. Preprint at https://arxiv.org/abs/2003.14395 (2020).
-
Luz, E. et al. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Preprint at http://arxiv.org/abs/2004.05717 (2020).
-
Bassi, P. R. A. S. & Attux, R. A deep convolutional neural network for COVID-19 detection using chest X-rays. Preprint at http://arxiv.org/abs/2005.01578 (2020).
-
Gueguim Kana, E. B., Zebaze Kana, M. G., Donfack Kana, A. F. & Azanfack Kenfack, R. H. A web-based diagnostic tool for COVID-19 using machine learning on chest radiographs (CXR). Preprint at medRxiv https://doi.org/10.1101/2020.04.21.20063263 (2020).
-
Heidari, M. et al. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inform. 144, 104284 (2020).
-
Li, X., Li, C. & Zhu, D. COVID-MobileXpert: on-device COVID-19 screening using snapshots of chest X-ray. Preprint at http://arxiv.org/abs/2004.03042 (2020).
-
Zokaeinikoo, M., Mitra, P., Kumara, S. & Kazemian, P. AIDCOV: an interpretable artificial intelligence model for detection of COVID-19 from chest radiography images. Preprint at medRxiv https://doi.org/10.1101/2020.05.24.20111922 (2020).
-
Sayyed, A. Q. M. S., Saha, D. & Hossain, A. R. CovMUNET: a multiple loss approach towards detection of COVID-19 from chest X-ray. Preprint at https://arxiv.org/abs/2007.14318 (2020).
-
Zhang, R. et al. COVID19XrayNet: a two-step transfer learning model for the COVID-19 detecting problem based on a limited number of chest X-ray images. Interdiscip. Sci. Comput. Life Sci. 12, 555–565 (2020).
-
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. In Proce. IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2818–2826 (IEEE Computer Society, 2016).
-
Tan, M. & Le, Q. EfficientNet: rethinking model scaling for convolutional neural networks. In International Conference of Machine Learning ICML 6105–6114 (PMLR, 2019).
-
Georgescu, B. et al. Machine learning automatically detects COVID-19 using chest CTs in a large multicenter cohort. Preprint at http://arxiv.org/abs/2006.04998 (2020).
-
Ko, H. et al. COVID-19 pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT image: model development and validation. J. Med. Internet Res. 22, e19569 (2020).
-
Wang, S. et al. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19). Preprint at medRxiv https://doi.org/10.1101/2020.02.14.20023028 (2020).
-
Pu, J. et al. Any unique image biomarkers associated with COVID-19? Eur. Radiol. https://doi.org/10.1007/s00330-020-06956-w (2020).
-
Amyar, A., Modzelewski, R. & Ruan, S. Multi-task deep learning based CT imaging analysis for COVID-19: classification and segmentation. Preprint at medRxiv https://doi.org/10.1101/2020.04.16.2006470 (2020).
-
Ardakani, A. A., Kanafi, A. R., Acharya, U. R., Khadem, N. & Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 121, 103795 (2020).
-
Mei, X. et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. https://doi.org/10.1038/s41591-020-0931-3 (2020).
-
Han, Z. et al. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans. Med. Imaging 39, 2584–2594 (2020).
-
Acar, E., Şahin, E. & Yilmaz, İ. Improving effectiveness of different deep learning-based models for detecting COVID-19 from computed tomography (CT) images. Preprint at medRxiv https://doi.org/10.1101/2020.06.12.20129643 (2020).
-
Chen, X., Yao, L., Zhou, T., Dong, J. & Zhang, Y. Momentum contrastive learning for few-shot COVID-19 diagnosis from chest CT images. Preprint at http://arxiv.org/abs/2006.13276 (2020).
-
Jin, S. et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. Preprint at medRxiv https://doi.org/10.1101/2020.03.19.20039354 (2020).
-
Wang, S. et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. https://doi.org/10.1183/13993003.00775-2020 (2020).
-
Shah, V. et al. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Preprint at medRxiv https://doi.org/10.1101/2020.07.11.20151332 (2020).
-
Wang, J. et al. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imaging 39, 2572–2583 (2020).
-
Wang, M. et al. Deep learning-based triage and analysis of lesion burden for COVID-19: a retrospective study with external validation. Lancet Digit. Health 2, e506–e515 (2020).
-
Deng, J. et al. ImageNet: a large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 (IEEE, 2010); https://doi.org/10.1109/cvpr.2009.5206848
-
Goodfellow, I. J. et al. Generative adversarial nets. In Proceedings of the 27th International Conference on Neural Information Processing Systems Vol. 2 2672–2680 (MIT Press, 2014).
-
Shi, F. et al. Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification. Preprint at http://arxiv.org/abs/2003.09860 (2020).
-
Guiot, J. et al. Development and validation of an automated radiomic CT signature for detecting COVID-19. Preprint at medRxiv https://doi.org/10.1101/2020.04.28.20082966 (2020).
-
Chen, X. X. et al. A diagnostic model for coronavirus disease 2019 (COVID-19) based on radiological semantic and clinical features: a multi-center study. Eur. Radiol. https://doi.org/10.1007/s00330-020-06829-2 (2020).
-
Qin, L. et al. A predictive model and scoring system combining clinical and CT characteristics for the diagnosis of COVID-19. Eur. Radiol. https://doi.org/10.1007/s00330-020-07022-1 (2020).
-
Xie, C. et al. Discrimination of pulmonary ground-glass opacity changes in COVID-19 and non-COVID-19 patients using CT radiomics analysis. Eur. J. Radiol. Open 7, 100271 (2020).
-
Xu, M. et al. Accurately differentiating COVID-19, other viral infection, and healthy individuals using multimodal features via late fusion learning. Preprint at medRxiv https://doi.org/10.1101/2020.08.18.20176776 (2020).
-
Chassagnon, G. et al. AI-driven CT-based quantification, staging and short-term outcome prediction of COVID-19 pneumonia. Preprint at medRxiv https://doi.org/10.1101/2020.04.17.20069187 (2020).
-
Ghosh, B. et al. A quantitative lung computed tomography image feature for multi-center severity assessment of COVID-19. Preprint at medRxiv https://doi.org/10.1101/2020.07.13.20152231 (2020).
-
Wei, W., Hu, X. W., Cheng, Q., Zhao, Y. M. & Ge, Y. Q. Identification of common and severe COVID-19: the value of CT texture analysis and correlation with clinical characteristics. Eur. Radiol. https://doi.org/10.1007/s00330-020-07012-3 (2020).
-
Li, M. D. et al. Improvement and multi-population generalizability of a deep learning-based chest radiograph severity score for COVID-19. Preprint at medRxiv https://doi.org/10.1101/2020.09.15.20195453 (2020).
-
Li, M. D. et al. Automated assessment and tracking of COVID-19 pulmonary disease severity on chest radiographs using convolutional siamese neural networks. Radiol. Artif. Intell. 2, e200079 (2020).
-
Schalekamp, S. et al. Model-based prediction of critical illness in hospitalized patients with COVID-19. Radiology https://doi.org/10.1148/radiol.2020202723 (2020).
-
Wang, X. et al. Multi-center study of temporal changes and prognostic value of a CT visual severity score in hospitalized patients with COVID-19. Am. J. Roentgenol. https://doi.org/10.2214/ajr.20.24044 (2020).
-
Yip, S. S. F. et al. Performance and robustness of machine learning-based radiomic COVID-19 severity prediction. Preprint at medRxiv https://doi.org/10.1101/2020.09.07.20189977 (2020).
-
Goncharov, M. et al. CT-based COVID-19 triage: deep multitask learning improves joint identification and severity quantification. Preprint at https://arxiv.org/abs/2006.01441 (2020).
-
Ramtohul, T. et al. Quantitative CT extent of lung damage in COVID-19 pneumonia is an independent risk factor for inpatient mortality in a population of cancer patients: a prospective study. Front. Oncol. 10, 1560 (2020).
-
Lassau, N. et al. AI-based multi-modal integration of clinical characteristics, lab tests and chest CTs improves COVID-19 outcome prediction of hospitalized patients. Preprint at medRxiv https://doi.org/10.1101/2020.05.14.20101972 (2020).
-
Yue, H. et al. Machine learning-based CT radiomics method for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: a multicenter study. Ann. Transl. Med. 8, 859–859 (2020).
-
Zhu, X. et al. Joint prediction and time estimation of COVID-19 developing severe symptoms using chest CT scan. Preprint at http://arxiv.org/abs/2005.03405 (2020).
-
Cohen, J. P. et al. Predicting COVID-19 pneumonia severity on chest X-ray with deep learning. Preprint at https://arxiv.org/abs/2005.11856 (2020).
-
Chao, H. et al. Integrative analysis for COVID-19 patient outcome prediction. Preprint at https://arxiv.org/abs/2007.10416 (2020).
-
Wu, Q. et al. Radiomics analysis of computed tomography helps predict poor prognostic outcome in COVID-19. Theranostics 10, 7231–7244 (2020).
-
Zheng, Y. et al. Development and validation of a prognostic nomogram based on clinical and CT features for adverse outcome prediction in patients with COVID-19. Korean J. Radiol. 21, 1007–1017 (2020).
-
Chen, Y. et al. A quantitative and radiomics approach to monitoring ards in COVID-19 patients based on chest CT: a retrospective cohort study. Int. J. Med. Sci. 17, 1773–1782 (2020).
-
Bai, H. X. et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other etiology on chest CT. Radiology 296, E156–E165 (2020).
-
Yang, X. et al. COVID-CT-Dataset: a CT scan dataset about COVID-19. Preprint at http://arxiv.org/abs/2003.13865 (2020).
-
Amyar, A., Modzelewski, R., Li, H. & Ruan, S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: classification and segmentation. Comput. Biol. Med. 126, 104037 (2020).
-
COVID-19 Radiography Database (Kaggle, accessed 29 July 2020); https://www.kaggle.com/tawsifurrahman/covid19-radiography-database
-
Cohen, J. P., Morrison, P. & Dao, L. COVID-19 image data collection. Preprint at http://arxiv.org/abs/2003.11597 (2020).
-
Kermany, D. S. et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172, 1122–1131.e9 (2018).
-
COVID-19: radiology reference article. Radiopaedia https://radiopaedia.org/articles/covid-19-4?lang=gb (accessed 29 July 2020).
-
COVID-19 Database (SIRM, accessed 29 July 2020); https://www.sirm.org/en/category/articles/covid-19-database/
-
CORONACASES.org (RAIOSS.com, accessed 30 July 2020); https://coronacases.org/
-
Eurorad (ESR, accessed 29 July 2020); https://www.eurorad.org/
-
Chest X-Ray Images (Pneumonia) (Kaggle, accessed 29 July 2020); https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
-
Maguolo, G. & Nanni, L. A critic evaluation of methods for COVID-19 automatic detection from X-ray images. Preprint at http://arxiv.org/abs/2004.12823 (2020).
-
RSNA Pneumonia Detection Challenge (Kaggle, accessed 29 July 2020); https://www.kaggle.com/c/rsna-pneumonia-detection-challenge
-
Bachtiger, P., Peters, N. & Walsh, S. L. Machine learning for COVID-19—asking the right questions. Lancet Digit. Health 2, e391–e392 (2020).
-
Banerjee, I. et al. Was there COVID-19 back in 2012? Challenge for AI in diagnosis with similar indications. Preprint at http://arxiv.org/abs/2006.13262 (2020).
-
Vickers, A. J. & Elkin, E. B. Decision curve analysis: a novel method for evaluating prediction models. Med. Decis. Making 26, 565–574 (2006).
-
Lambin, P. et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 14, 749–762 (2017).
-
Mongan, J., Moy, L. & Kahn, C. E. Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiol. Artif. Intell. 2, e200029 (2020).
-
Collins, G. S., Reitsma, J. B., Altman, D. G. & Moons, K. G. M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Ann. Intern. Med. 162, 55–63 (2015).
-
Wolff, R. F. et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann. Intern. Med. 170, 51 (2019).
-
Whiting, P. F. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 155, 529 (2011).
-
Zhang, C., Bengio, S., Hardt, M., Recht, B. & Vinyals, O. Understanding deep learning requires rethinking generalization. In 5th International Conference on Learning Representations ICLR 2017 (ICLR, 2017).
-
Liberati, A. et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 339, b2700 (2009).
-
COVID-19 Open Access Project: Living Evidence on COVID-19 (accessed 21 January 2021, COVID-19 Open Access Project); https://ispmbern.github.io/covid-19/living-review/
-
Pneumonia of unknown cause—China. WHO https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/ (2020).
-
Covidence Systematic Review Software v2486 6284093b (Veritas Health Innovation, 2020); https://www.covidence.org/
-
Aviles-Rivero, A. I., Sellars, P., Schönlieb, C.-B. & Papadakis, N. GraphXCOVID: explainable deep graph diffusion pseudo-labelling for identifying COVID-19 on chest X-rays. Preprint at https://arxiv.org/abs/2010.00378 (2020).
Acknowledgements
M.R. acknowledges the support of his supervisors M. Patel and A. Reynolds at AstraZeneca. The AIX-COVNET collaboration also acknowledges the support of Intel in supporting the collaboration. There is no direct funding source for this study; however, the following authors do report indirect funding as follows: AstraZeneca (M.R.), European Research Council under the European Union’s Horizon 2020 research and innovation programme grant agreement no. 777826 (M.T.), Wellcome Trust (C.E, C.M., J.H.F.R.), The Mark Foundation for Cancer Research (L.B., E.S.), Cancer Research UK Cambridge Centre (C9685/A25177) (L.B., E.S., C.-B.S.), British Heart Foundation (Z.T., J.H.F.R.), the NIHR Cambridge Biomedical Research Centre (Z.T., J.H.F.R.), HEFCE (J.H.F.R.), Lyzeum Ltd. (J.G.), The Gates Cambridge Trust (D.D.), the Cambridge International Trust (S.U.), the EPSRC Cambridge Mathematics of Information in Healthcare Hub EP/T017961/1 (A.I.A.-R., J.H.F.R., C.-B.S.) and the Cantab Capital Institute for the Mathematics of Information (A.I.A.-R, C.-B.S.). In addition, C.-B.S. acknowledges support from the Leverhulme Trust project on ‘Breaking the non-convexity barrier’, the Philip Leverhulme Prize, the EPSRC grants EP/S026045/1 and EP/T003553/1, the Wellcome Innovator Award RG98755, European Union Horizon 2020 research and innovation programmes under the Marie Skłodowska-Curie grant agreement no. 777826 NoMADS and no. 691070 CHiPS and the Alan Turing Institute. All authors had full access to all the data in the study and the corresponding author had final responsibility for the decision to submit for publication.
Author information
Author notes
- These authors contributed equally: James H. F. Rudd, Evis Sala, Carola-Bibiane Schönlieb.
Affiliations
- Department of Applied Mathematics and Theoretical Physics, University of Cambridge, Cambridge, UKMichael Roberts, Derek Driggs, Julian Gilbey, Angelica I. Aviles-Rivero, Christian Etmann, Jan Stanczuk & Carola-Bibiane Schönlieb
- Oncology R&D, AstraZeneca, Cambridge, UKMichael Roberts
- Department of Mathematics, University of Manchester, Manchester, UKMatthew Thorpe
- Department of Radiology, University of Cambridge, Cambridge, UKMichael Yeung, Stephan Ursprung, Cathal McCague, Lucian Beer, Jonathan R. Weir-McCall, Zhongzhao Teng, Lorena Escudero Sánchez, Maria Delgado Ortet & Evis Sala
- Cancer Research UK Cambridge Centre, University of Cambridge, Cambridge, UKStephan Ursprung, Cathal McCague, Lorena Escudero Sánchez, Paula Martin Gonzalez & Evis Sala
- Royal Papworth Hospital, Cambridge, Royal Papworth Hospital NHS Foundation Trust, Cambridge, UKJonathan R. Weir-McCall & Alessandro Ruggiero
- Department of Infectious Diseases, Cambridge University Hospitals NHS Trust, Cambridge, UKEffrossyni Gkrania-Klotsas
- Department of Medicine, University of Cambridge, Cambridge, UKMuhunthan Thillai & James H. F. Rudd
- Qureight Ltd, Cambridge, UKAlessandro Ruggiero & Muhunthan Thillai
- Language Technology Laboratory, University of Cambridge, Cambridge, UKAnna Korhonen
- Population Health and Genomics, School of Medicine, University of Dundee, Dundee, UKEmily Jefferson
- Chelsea and Westminster NHS Trust and Royal Brompton NHS Hospital, London, UKEmmanuel Ako
- Department of Biomedical Imaging and Image-guided Therapy, Computational Imaging Research Lab Medical University of Vienna, Vienna, AustriaGeorg Langs, Helmut Prosch & Johannes Hofmanninger
- Department of Physics, University of Helsinki, Helsinki, FinlandGhassem Gozaliasl
- National Heart and Lung Institute, Imperial College London, London, UKGuang Yang
- Boehringer, Ingelheim, GermanyHelmut Prosch
- Addenbrooke’s Hospital, Cambridge University Hospitals NHS Trust, Cambridge, UKJacobus Preller & Judith Babar
- Research Program in System Oncology, Faculty of Medicine, University of Helsinki, Helsinki, FinlandJing Tang & Tolou Shadbahr
- Interstitial Lung Disease Unit, Royal Papworth Hospital, Cambridge, Royal Papworth Hospital NHS Foundation Trust, Cambridge, UKMuhunthan Thillai
- Biopharmaceuticals R&D, AstraZeneca, Cambridge, UKPhilip Teare & Mishal Patel
- Signal Processing in Earth Observation, Technical University of Munich, Munich, GermanyXiaoxiang Zhu
- Department of Chemistry, University of Cambridge, Cambridge, UKConor Cafolla
- AINOSTICS Ltd, Manchester, UKHojjat Azadbakht
- Centre for Medical Image Computing, University College London, London, UKJoseph Jacob
- SparkBeyond UK Ltd, London, UKJosh Lowe & Kyle Bradley
- Center for Biomedicine and Innovations at Faculty of Medicine, Macau University of Science and Technology, Macau, ChinaKang Zhang
- contextflow GmbH, Vienna, AustriaMarcel Wassin & Markus Holzer
- Cavendish Laboratory, University of Cambridge, Cambridge, UKKangyu Ji
- Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, ChinaTao Ai
- Institute of Astronomy, University of Cambridge, Cambridge, UKNicholas Walton
- Department of Computer Science and Technology, University of Cambridge, Cambridge, UKPietro Lio
- Department of Chemical Engineering and Biotechnology, University of Cambridge, Cambridge, UKSamuel Stranks
- Department of Engineering, University of Cambridge, Cambridge, UKWeizhe Lin
- Department of Radiology, Renmin Hospital of Wuhan University, Wuhan, ChinaYunfei Zha
- Aladdin Healthcare Technologies Ltd, London, UKZhangming Niu
Consortia
AIX-COVNET
- Michael Roberts
- , Derek Driggs
- , Matthew Thorpe
- , Julian Gilbey
- , Michael Yeung
- , Stephan Ursprung
- , Angelica I. Aviles-Rivero
- , Christian Etmann
- , Cathal McCague
- , Lucian Beer
- , Jonathan R. Weir-McCall
- , Zhongzhao Teng
- , Effrossyni Gkrania-Klotsas
- , Alessandro Ruggiero
- , Anna Korhonen
- , Emily Jefferson
- , Emmanuel Ako
- , Georg Langs
- , Ghassem Gozaliasl
- , Guang Yang
- , Helmut Prosch
- , Jacobus Preller
- , Jan Stanczuk
- , Jing Tang
- , Johannes Hofmanninger
- , Judith Babar
- , Lorena Escudero Sánchez
- , Muhunthan Thillai
- , Paula Martin Gonzalez
- , Philip Teare
- , Xiaoxiang Zhu
- , Mishal Patel
- , Conor Cafolla
- , Hojjat Azadbakht
- , Joseph Jacob
- , Josh Lowe
- , Kang Zhang
- , Kyle Bradley
- , Marcel Wassin
- , Markus Holzer
- , Kangyu Ji
- , Maria Delgado Ortet
- , Tao Ai
- , Nicholas Walton
- , Pietro Lio
- , Samuel Stranks
- , Tolou Shadbahr
- , Weizhe Lin
- , Yunfei Zha
- , Zhangming Niu
- , James H. F. Rudd
- , Evis Sala
- & Carola-Bibiane Schönlieb
Contributions
M.R., D.D., M.T., J.G., M.Y., A.I.A.-R., C.E., C.M., S.U. and L.B. contributed to the literature search, screening and quality assessment. M.R., D.D., M.T., J.G., M.Y., A.I.A.-R., C.E., C.M. and S.U. contributed to the data extraction, analysis and interpretation along with writing the original draft manuscript. M.R., D.D., M.T., J.G., M.Y., A.I.A.-R., C.E., C.M., S.U., L.B., J.R.W.-M., Z.T., E.G.-K., J.H.F.R., E.S. and C.-B.S. contributed critical revisions of the manuscript, and all authors approved the final manuscript.
Corresponding author
Roberts, M., Driggs, D., Thorpe, M. et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell 3, 199–217 (2021). https://doi.org/10.1038/s42256-021-00307-0
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks Pengtao Xie and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Discussions 1 and 2 and Table 1.
Supplementary Data 1
Workbook detailing all papers considered during each stage of the screening.
Supplementary Data 2
Workbook containing all of the CLAIM, RQS and PROBAST results.
Open Access
This article has been reproduced in accordance with its Creative Commons Attribution 4.0 International License, a copy of this license can be viewed at http://creativecommons.org/licenses/by/4.0/. The original source for this article can be found here.
